X-Ray Crystallography
X-ray crystallography (XRC) is the experimental technique of determining the atomic and molecular structure of a crystal, in which the crystalline structure scatter into many specific directions or diffract, a beam of incident X-rays. By measuring the angles and intensities of these diffracted beams, a three-dimensional picture of the density of electrons within the crystal can be produced. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, crystallographic disorder and other various information[1].
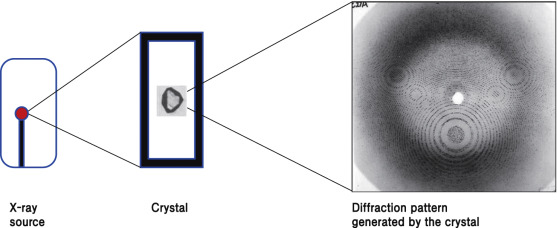
Figure: Principle of X-ray crystallography. High-resolution structure can be obtained from the diffraction pattern generated by crystals[2].
Many materials can form crystals such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules, which was instrumental in X-ray crystallography becoming fundamental in the development of many scientific fields. This technique helps to determine the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among various materials, especially minerals and alloys. The method also established the structure and function of many biological molecules, including vitamins, drugs, proteins and nucleic acids such as DNA. X-ray crystallography is still the primary method for characterization of the atomic structure of new materials.
In a single-crystal X-ray diffraction measurement, a crystal is placed on a goniometer. The goniometer is used to position the crystal at selected orientations. The crystal is illuminated with a finely focused monochromatic beam of X-rays, producing a diffraction pattern of regularly spaced spots known as reflections. 2D images taken at different orientations are converted into a 3D model of the density of electrons within the crystal, using the mathematical method of Fourier transforms, combined with chemical data known for the sample. Poor resolution (fuzziness) or even errors may result if the crystals are too small, or not uniform enough.
If single crystals of sufficient size cannot be obtained, various other X-ray methods can be applied to obtain less detailed information. Such methods include fiber diffraction, powder diffraction and (if the sample is not crystallized) small-angle X-ray scattering (SAXS). If the material is only available in the form of nanocrystalline powders or suffers from poor crystallinity, the methods of electron crystallography can be applied for determining the atomic structure. For all these mentioned X-ray diffraction methods, the scattering is elastic. The scattered X-rays have the same wavelength as the incoming X-ray. However, inelastic X-ray scattering methods are useful in studying excitations of the sample such as plasmons, crystal-field and orbital excitations, magnons, and phonons, rather than the distribution of its atoms.
X-ray crystallography is one of the main tools to solve crystal structures of proteins, nucleic acids and other biological molecules[3]. Also it is routinely used to determine how a pharmaceutical drug interacts with its protein target and what changes might improve it [4]. However, intrinsic membrane proteins remain challenging to crystallize because they require detergents or other denaturants to solubilize them in isolation. Such detergents often interfere with crystallization. Membrane proteins are a large component of the genome, and include many proteins of great physiological importance, such as ion channels and receptors[5,6]. Helium cryogenics are used to prevent radiation damage in protein crystals.[7]
Despite being a valuable tool in structural biology, protein crystallography has some problems in its methodology, that hinder data interpretation. The crystal lattice, which is formed during the crystallization process, contains numerous units of the purified protein, which are densely and symmetrically packed in the crystal. When looking for a previously unknown protein, figuring out its shape and boundaries within the crystal lattice can be challenging. Proteins are usually composed of smaller subunits, and the task of distinguishing between the subunits and identifying the actual protein, can be challenging even for the experienced crystallographers. The non-biological interfaces that occur during crystallization are known as crystal-packing contacts (or simply, crystal contacts) and cannot be distinguished by crystallographic means. When a new protein structure is solved by X-ray crystallography and deposited in the Protein Data Bank, its authors are requested to specify the “biological assembly” which would constitute the functional, biologically-relevant protein. However, errors, missing data and inaccurate annotations during the submission of the data, give rise to obscure structures and compromise the reliability of the database. The error rate in the case of faulty annotations alone has been reported to be upwards of 6.6%[8] or approximately 15%,[9] arguably a non-trivial size considering the number of deposited structures. This “interface classification problem” is typically tackled by computational approaches and has become a recognized subject in structural bioinformatics.
References:
- “Resonant X-ray Scattering | Shen Laboratory”. arpes.stanford.edu. Retrieved 2019-07-10
- Wang-Shick Ryu, (2017), Molecular Virology of Human Pathogenic Viruses, pp.21-29
- “Table of entries in the PDB, arranged by experimental method” https://www.rcsb.org/pdb/statistics/holdings.do
- Scapin G (2006). “Structural biology and drug discovery”. Current Pharmaceutical Design. 12 (17): 2087–97. doi:2174/138161206777585201. PMID 16796557
- Lundstrom, K., (November 2006). “Structural genomics for membrane proteins”. Cellular and Molecular Life Sciences. 63(22): 2597–607. doi:1007/s00018-006-6252-y. PMID 17013556. S2CID 13432321.
- Lundstrom, K., (August 2004). “Structural genomics on membrane proteins: mini review”. Combinatorial Chemistry & High Throughput Screening. 7(5): 431–9. doi:2174/1386207043328634. PMID 15320710.
- Chinte, U, Shah, B, Chen, Y.S., Pinkerton AA, Schall CA, Hanson BL (April 2007). “Cryogenic (<20 K) helium cooling mitigates radiation damage to protein crystals”. Acta Crystallographica. Section D, Biological Crystallography. 63(Pt 4): 486–92. doi:1107/s0907444907005264. PMID 17372353.
- Baskaran K, Duarte JM, Biyani N, Bliven S, Capitani G (October 2014). “A PDB-wide, evolution-based assessment of protein-protein interfaces”. BMC Structural Biology. 14 (1): 22. doi:1186/s12900-014-0022-0. PMC 4274722. PMID 25326082.
- Levy ED (November 2007). “PiQSi: protein quaternary structure investigation”. Structure. 15 (11): 1364–7. doi:1016/j.str.2007.09.019. PMID17997962.